This past month has kept me busy, which would explain the large gap between posts!
The highlight, if you want to call it that, was my first real job interview. It was nerve-racking, but I did a good deal of prep leading up to the interview and I think it went okay. Crazy that there’s only two months left in my practicum… then I’ll be feverishly studying for the CSMLS exam and hopefully entering the workforce!
Microbiology is the longest of all our rotations at 10 weeks. The last few benches I’ve been on were Respiratory, Genital, and Deep Wounds. Each site can be branched even further—respiratory specimens include those from the nose, throat, lower respiratory tract (e.g. sputum), and less invasive ear and eye collections.
Specimens are set up to plates and/or other media suitable for the identification of pathogens, and potential pathogens, from that specific site. ‘Potential pathogen’ is used to describe organisms which are of significance only when they meet specific criteria – often dictated by their amount(s) in relation to normal microbial flora, and the number of white blood cells seen in the Gram stain. White cells fight infection, hence an increased count could suggest pathogenic cause.
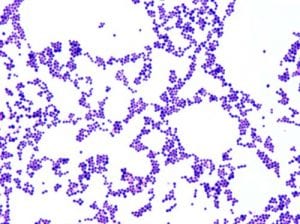
Nasal swabs are processed only to detect carriage of Staphylococcus aureus. S. aureus is a gram positive cocci and one of the most commonly isolated pathogens in the lab. Colonization with the bacteria may predispose recurrent skin and/or wound infections following surgery; a nasal swab may be collected to screen pre-operative and ER patients.
Are you a BCIT News insider? Sign-up to receive the latest news on BCIT.
From my perspective, there are two major components to Microbiology: the work-up and identification of clinically significant organisms, and antimicrobial susceptibility testing (AST). AST tests a number of antimicrobial agents (drugs) against an organism to identify which would be most effective in treating the infection.
Not all clinically significant organisms undergo AST. Those with a predictable susceptibility pattern can be treated empirically—penicillin is used to treat pharyngitis caused by Group A streptococci, as the organism has no known resistance to the drug.
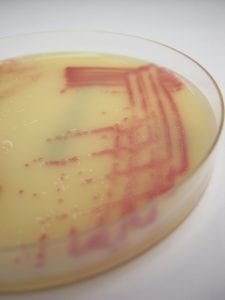
Methicillin-resistant S. aureus (MRSA) strains are most commonly seen in hospital and health care settings, but are becoming more frequent in the community population. Why does this matter? Because the notion has been that community-acquired infections are generally easier to treat than their hospital-acquired counterparts; increasing drug resistance hinders the accessibility of more common antibiotics.
Nasal swabs can be cultured to MRSA selective agar, with MRSA appearing as pink colonies. Non-MRSA organisms may be white, colourless, or inhibited.
A couple other fast facts about MRSA—their resistance is due to an altered penicillin binding protein bound to the bacteria’s cell membrane. The altered target site has a lower binding affinity for beta-lactam drugs, rendering them ineffective.
MRSA strains are also hetero-resistant, meaning they have both antimicrobial-susceptible and antimicrobial-resistant subpopulations.
Interested in the Medical Laboratory Science full-time diploma program? Join the next online info session at BCIT.

Hi
Could you tell me which score is necessary for lab technologies?